Introduction: Anaplastic large cell lymphomas (ALCLs) are CD30-positive T-cell lymphomas classified into ALK-positive and ALK-negative. The ALK-negative ALCLs are a genetically heterogeneous group that includes DUSP22-rearranged, TP63-rearranged and triple-negative cases. The DUSP22-rearranged ALCLs characteristically grow in a diffuse sheet-like pattern and are composed of intermediate-sized monomorphous hallmark cells, admixed with smaller cells with prominent nuclear indentations and occasional central nuclear pseudo inclusions (King RL et al, Am J Surg Pathol. 2016). Further, DUSP22-rearranged ALCL is a molecularly distinct subset of ALCL with a unique immunogenic phenotype that portends a favorable prognosis (Luchtel RA et al. Blood 2018).
Lymphoid enhancer binding factor (LEF1) is a crucial nuclear mediator of the Wnt/ β-catenin pathway and a transcription factor involved in T-cell maturation. However, limited data exists on the association of LEF1 in T-cell lymphomas, including ALCL. The aims of this study were to explore LEF1 expression by immunohistochemistry in different subsets of ALCLs and to analyze the association of Wnt/ β-catenin pathway in this cohort.
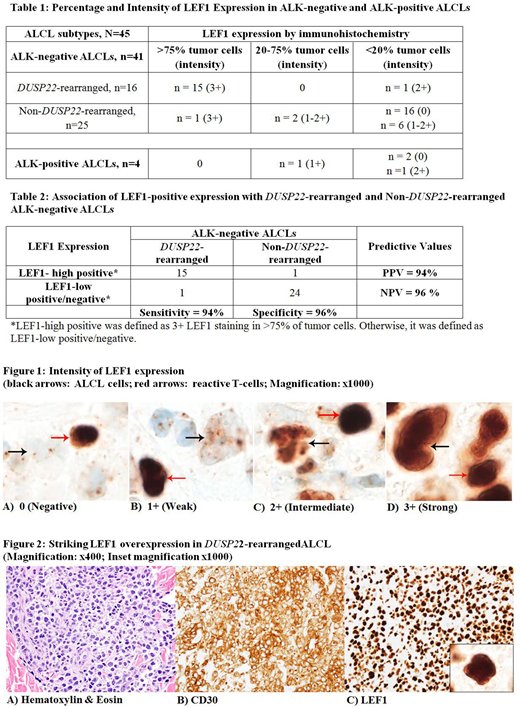
Methods: After approval by the institutional review board, we screened pathology reports from January 1, 1995 to December 31, 2019 and identified patients with a diagnosis of ALCL using our institutional database. Details on fluorescence in situ hybridization studies in ALK-negative cases were documented. Immunostaining for LEF1 and β catenin was performed on all cases where formalin fixed paraffin embedded tissue was available. The presence or absence of β catenin nuclear staining was recorded. LEF1 nuclear staining in the neoplastic ALCL cells was graded as negative (0), weak (1+), intermediate (2+) or strong (3+) and the percentage of LEF1-positive neoplastic cells was detailed (Figure 1). LEF1 scoring was performed by manual counting of the stained nuclei out of all tumor nuclei by two pathologists (A.R. and M.S.) and the grading was reviewed by another pathologist (P.J.K.).
Results: We evaluated 45 ALCL cases, including 41 (91.1%) ALK-negative and 4 (8.9%) ALK-positive ALCLs. Among the ALK-negative ALCLs, there were16 DUSP22-rearranged, and 25 non-DUSP22-rearanged (7 TP63-rearranged and 18 triple-negative) cases. Fifteen (94%) of 16 DUSP22-rearranged ALCLs showed strong expression of LEF1 in >75% tumor cells (Figure 2). In contrast, this strong and uniform LEF1 overexpression was only detected in 1 of 29 (3%) of the remaining ALCL cases (P <.0001) (Table 1).
We previously characterized a distinctive molecular signature in ALK-negative ALCLs with DUSP22-rearrangements using gene expression microarray analysis on frozen ALCL tissue samples (Luchtel RA et al. Blood 2018). On examining LEF1 gene expression in this previously published dataset, DUSP22 -rearranged cases had significantly higher LEF1 expression than any other genetic subtype (triple-negative, TP63-rearranged, or ALK-positive). Overall, mean LEF1 expression was 6.3-fold higher in ALCLs with DUSP22 rearrangements than in ALCLs without DUSP22-rearrangements (P=0.0001).
LEF1 is a coactivator of the Wnt/β-catenin pathway; therefore, it is reasonable to hypothesize the overexpression of LEF1 may promote Wnt/β-catenin signaling. However, all LEF1-positive cases were negative for β-catenin by immunohistochemistry, suggesting that LEF1 overexpression is independent of the Wnt/β-catenin pathway signaling in these cases.
Conclusions: There is a striking association between high LEF1 expression and DUSP22-rearrangements in ALCLs. The strong and uniform LEF1 expression pattern has a high positive predictive value (94%) and high negative predictive value (96%) for DUSP22- rearrangement in ALK-negative ALCLs (Table 2). The combination of characteristic morphologic and molecular features of DUSP22-rearranged cases with the unique LEF1 expression further emphasizes that DUSP22-rearranged ALCL represents a distinct clinicopathologic subset of ALCL.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal